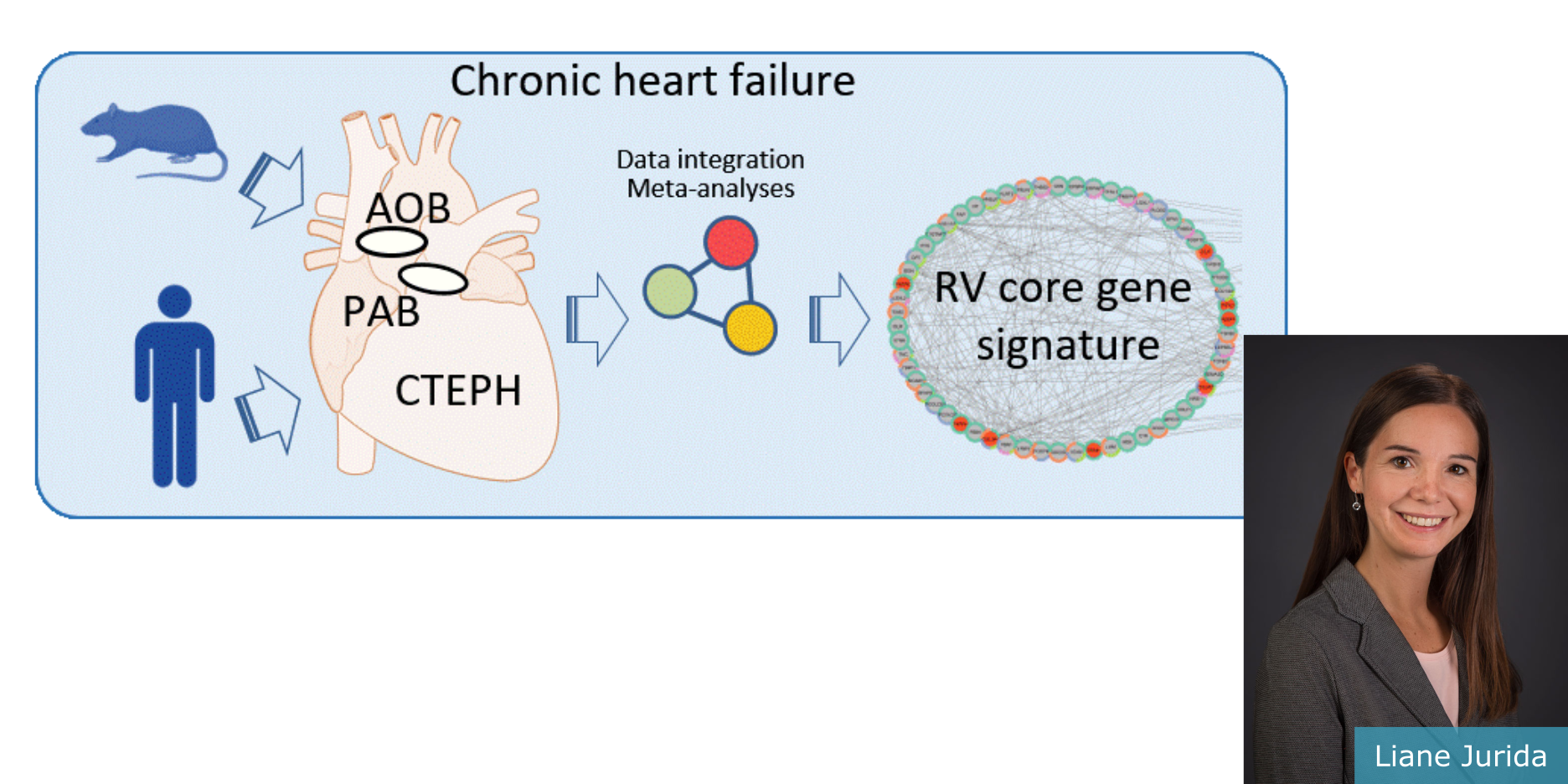
The molecular mechanisms of progressive right heart failure are incompletely understood. Jurida et al. identified more than 50 genes whose expression levels correlated with the severity of right heart disease.
In this study, Jurida et al. systematically examined transcriptomic changes occurring over months in isolated cardiomyocytes or whole heart tissues from failing right and left ventricles in rat models of pulmonary artery banding (PAB) or aortic banding (AOB).
Detailed bioinformatics analyses resulted in the identification of gene signature, protein and transcription factor networks specific to ventricles and compensated or decompensated disease states. Proteomic and RNA-FISH analyses confirmed PAB-mediated regulation of key genes and revealed spatially heterogeneous mRNA expression in the heart.
Intersection of rat PAB-specific gene sets with transcriptome datasets from human patients with chronic thromboembolic pulmonary hypertension (CTEPH) led to the identification of more than 50 genes whose expression levels correlated with the severity of right heart disease, including multiple matrix-regulating and secreted factors.
According to comparisons with published single cell (sc) / single nucleus (sn) RNA-seq data from humans with cardiomyopathies, several of these genes are likely to become activated in specific cardiac niches. These data define a conserved, differentially regulated genetic network associated with right heart failure in rats and humans.
Find the full article here:
Source: July 2024 (cpi-online.de)